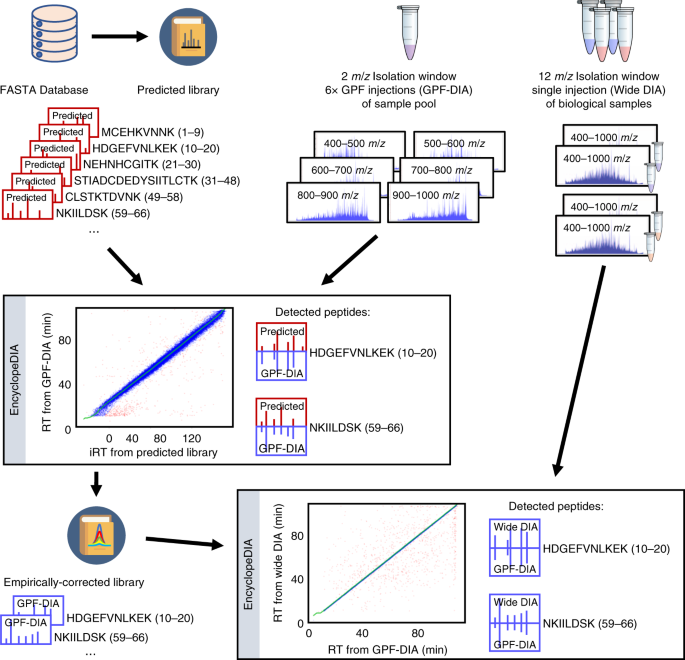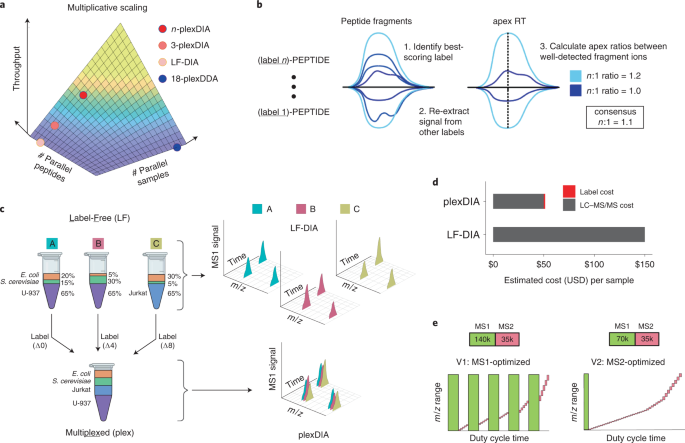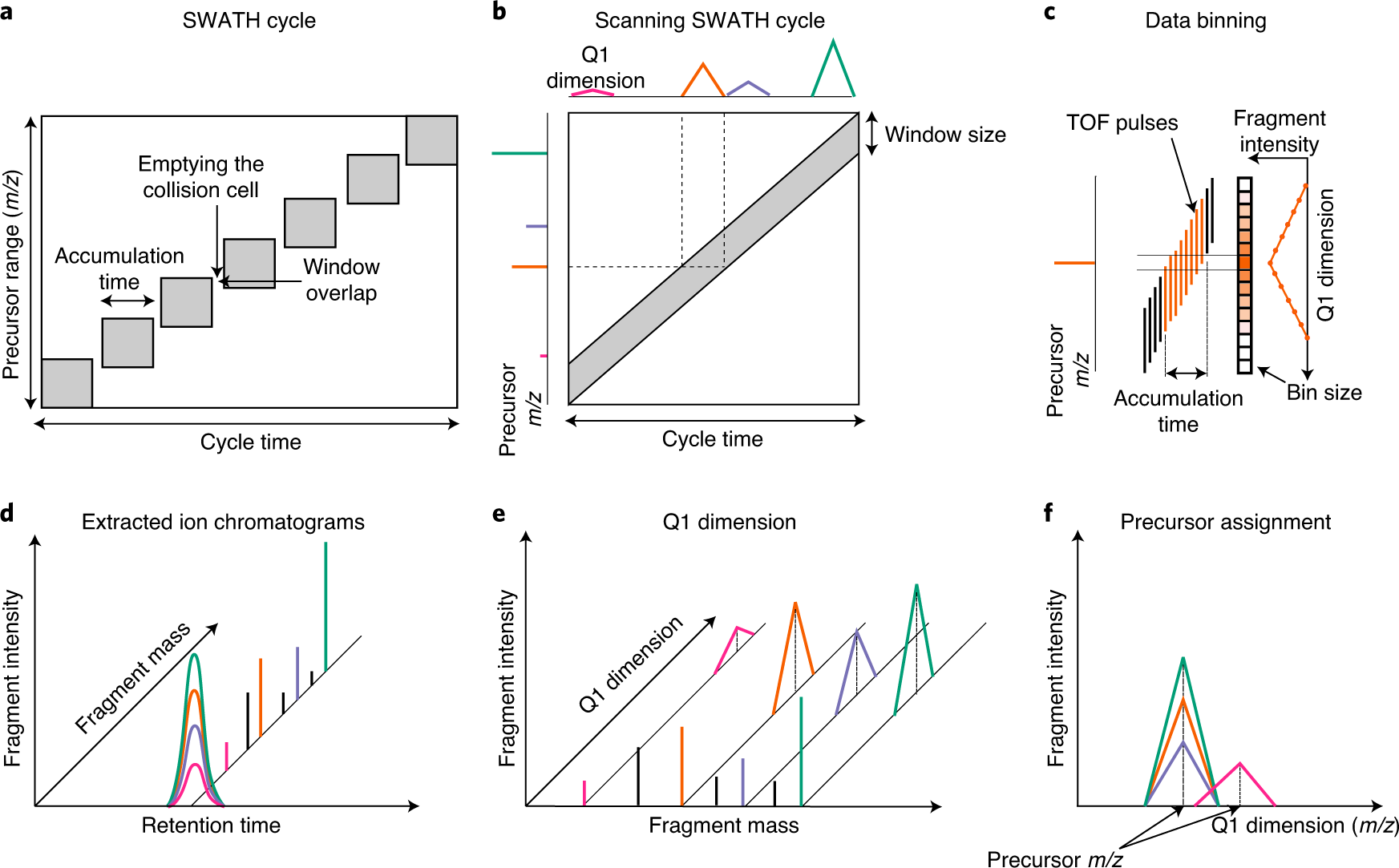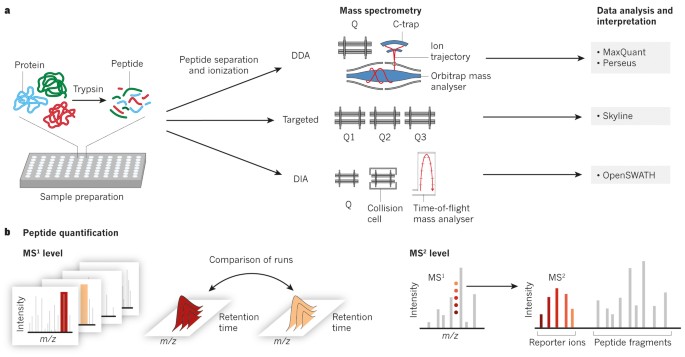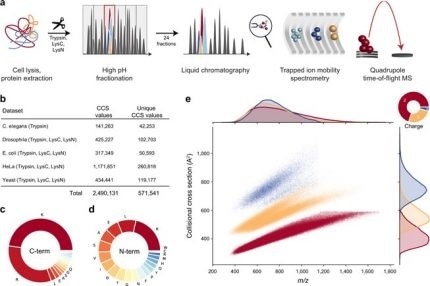
New Nature Communications publication by Mann & Theis Groups harnesses the benefits of large-scale peptide
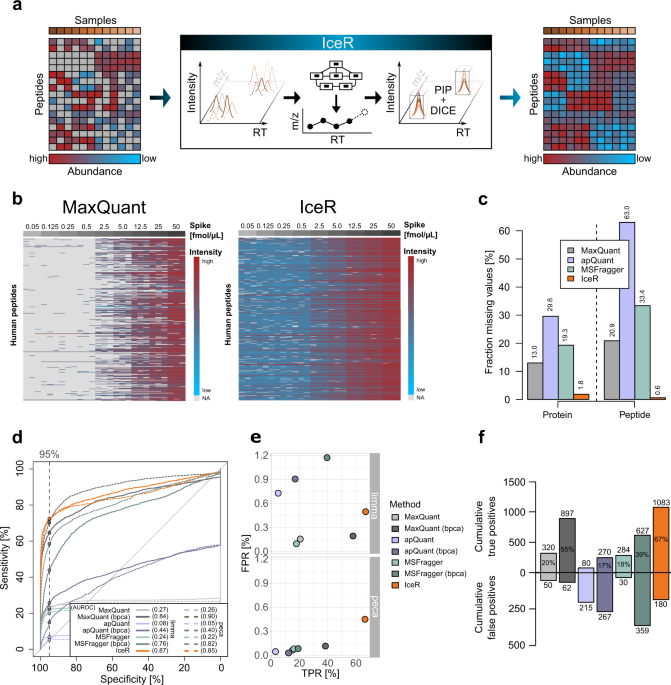
IceR improves proteome coverage and data completeness in global and single-cell proteomics | Nature Communications
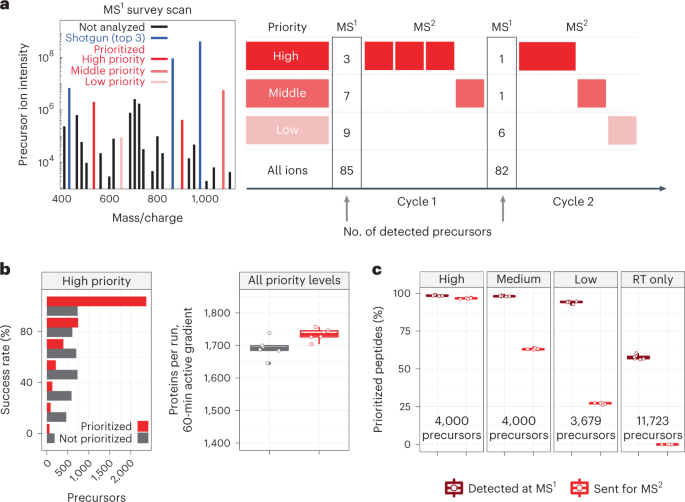
Prioritized mass spectrometry increases the depth, sensitivity and data completeness of single-cell proteomics | Nature Methods
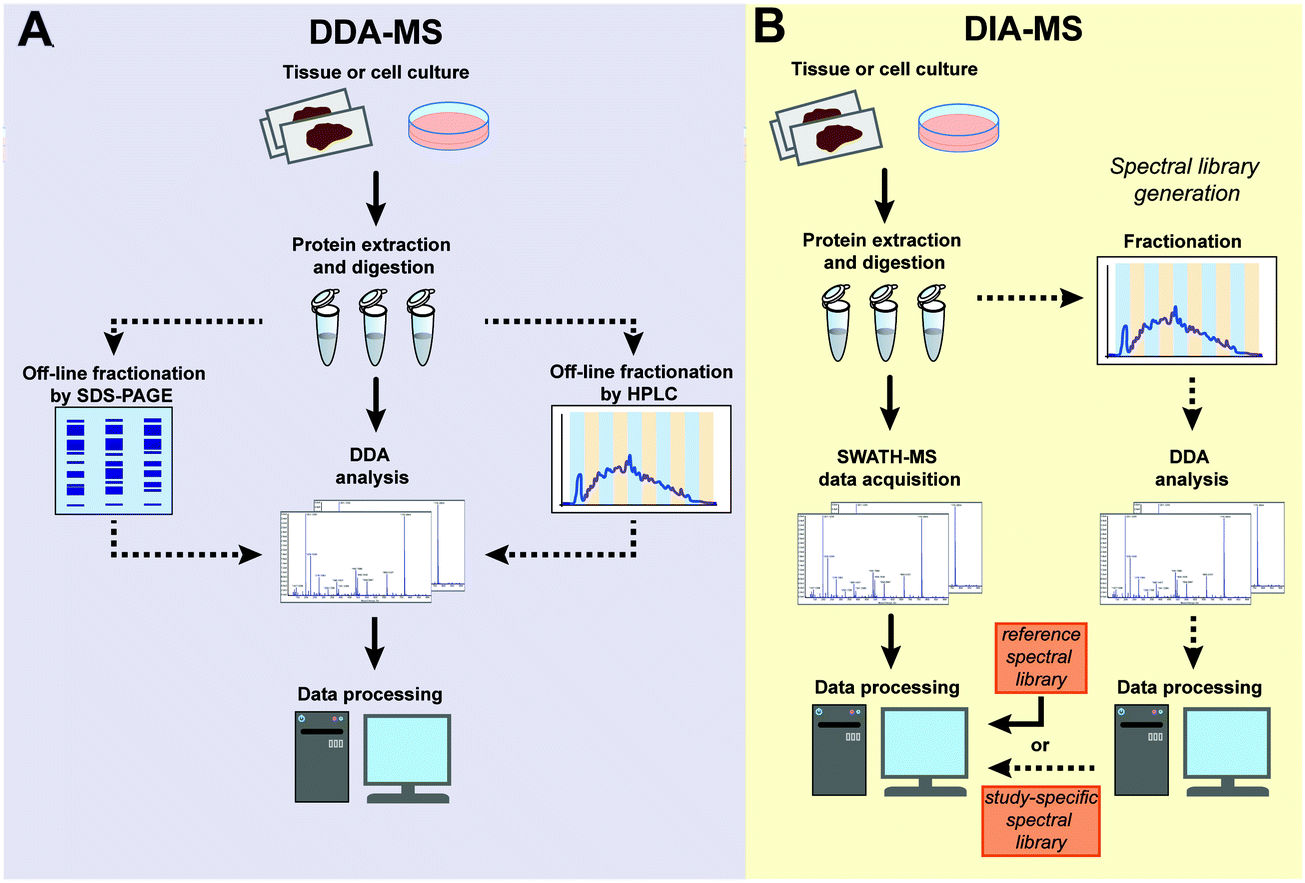
Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology - Molecular Omics (RSC Publishing) DOI:10.1039/D0MO00072H

Group-DIA: analyzing multiple data-independent acquisition mass spectrometry data files | Nature Methods

Hybrid-DIA: intelligent data acquisition integrates targeted and discovery proteomics to analyze phospho-signaling in single spheroids | Nature Communications
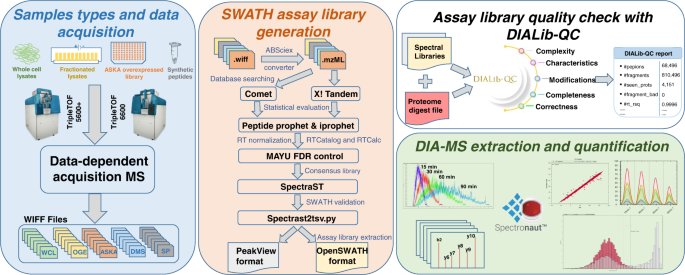
A comprehensive spectral assay library to quantify the Escherichia coli proteome by DIA/SWATH-MS | Scientific Data
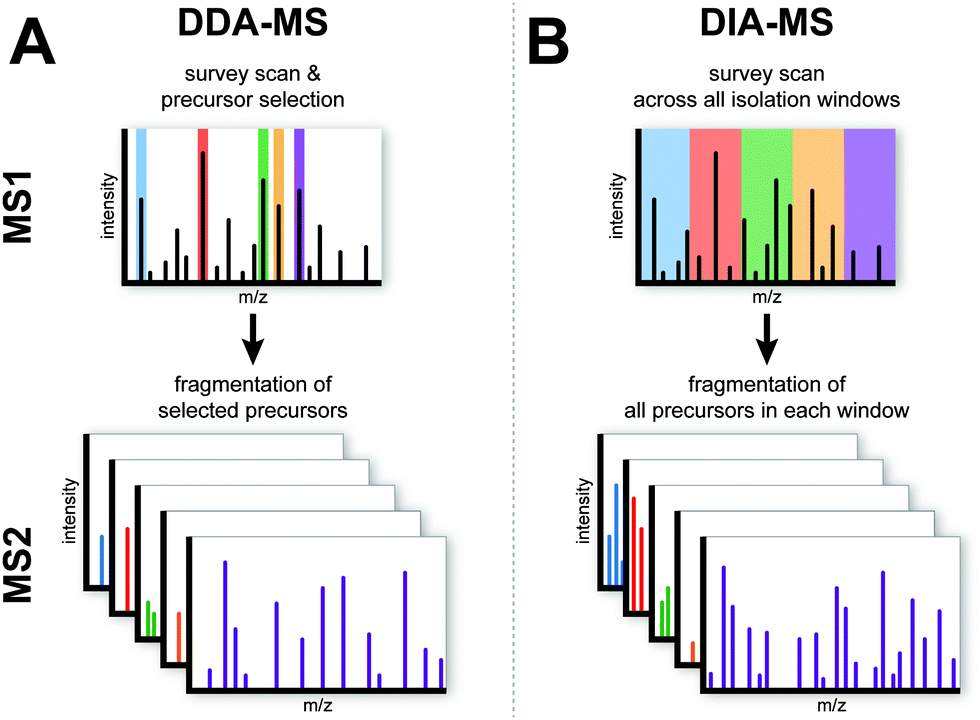
Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology - Molecular Omics (RSC Publishing) DOI:10.1039/D0MO00072H

Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry | Nature Communications

Use of Hybrid Data-Dependent and -Independent Acquisition Spectral Libraries Empowers Dual-Proteome Profiling | Journal of Proteome Research
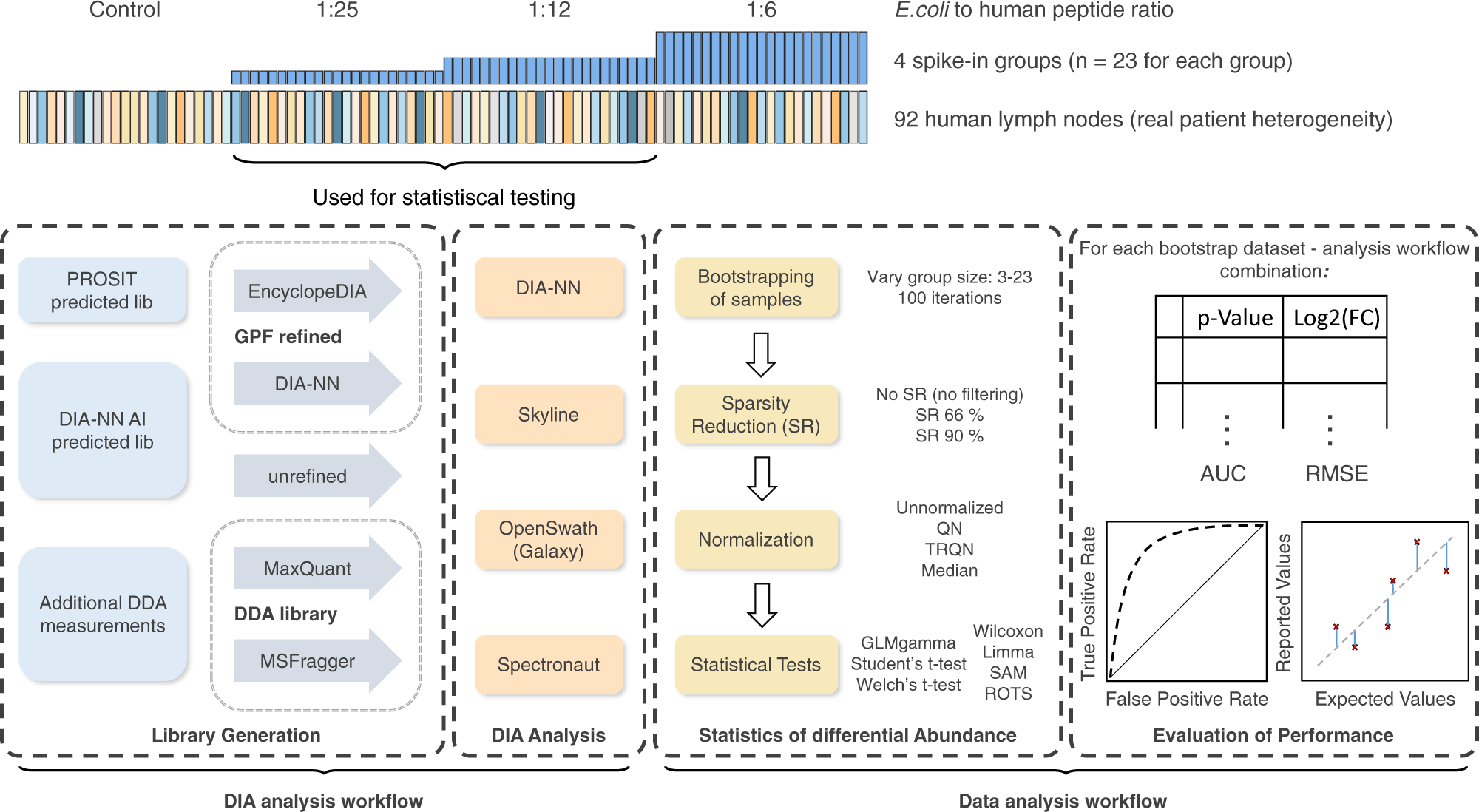
Benchmarking of analysis strategies for data-independent acquisition proteomics using a large-scale dataset comprising inter-patient heterogeneity | Nature Communications

Data-independent acquisition method for ubiquitinome analysis reveals regulation of circadian biology | Nature Communications
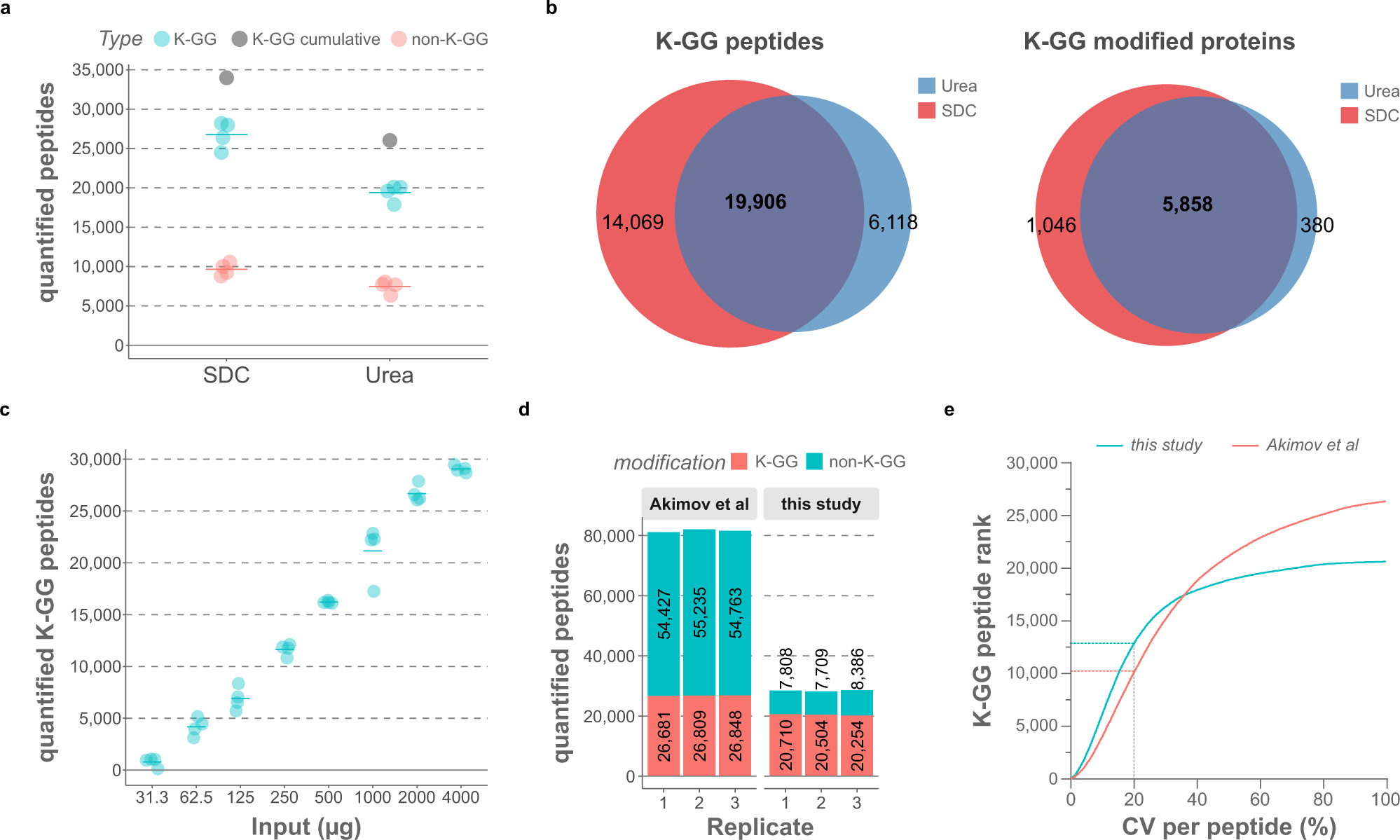
Time-resolved in vivo ubiquitinome profiling by DIA-MS reveals USP7 targets on a proteome-wide scale | Nature Communications

Deep Proteomics Using Two Dimensional Data Independent Acquisition Mass Spectrometry | Analytical Chemistry

Implementing the reuse of public DIA proteomics datasets: from the PRIDE database to Expression Atlas | Scientific Data
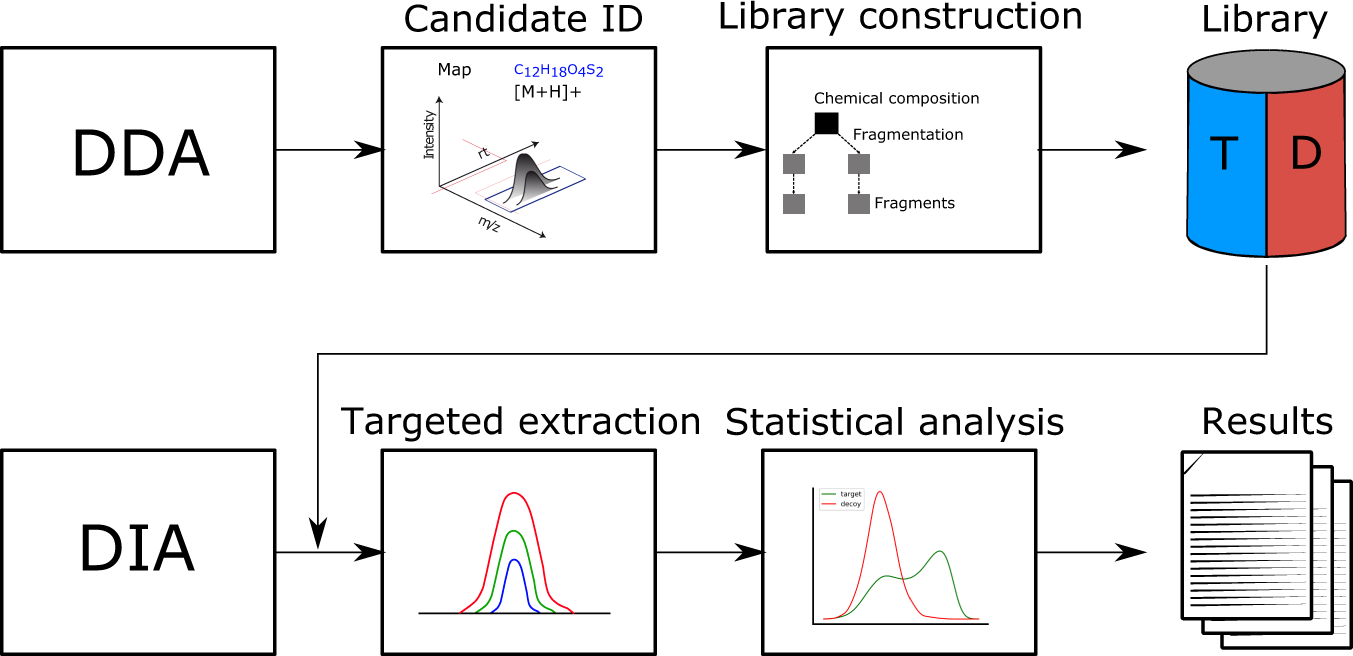
DIAMetAlyzer allows automated false-discovery rate-controlled analysis for data-independent acquisition in metabolomics | Nature Communications
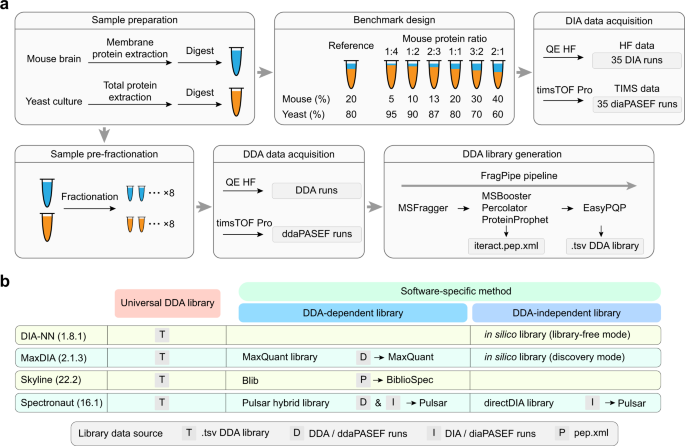
Benchmarking commonly used software suites and analysis workflows for DIA proteomics and phosphoproteomics | Nature Communications
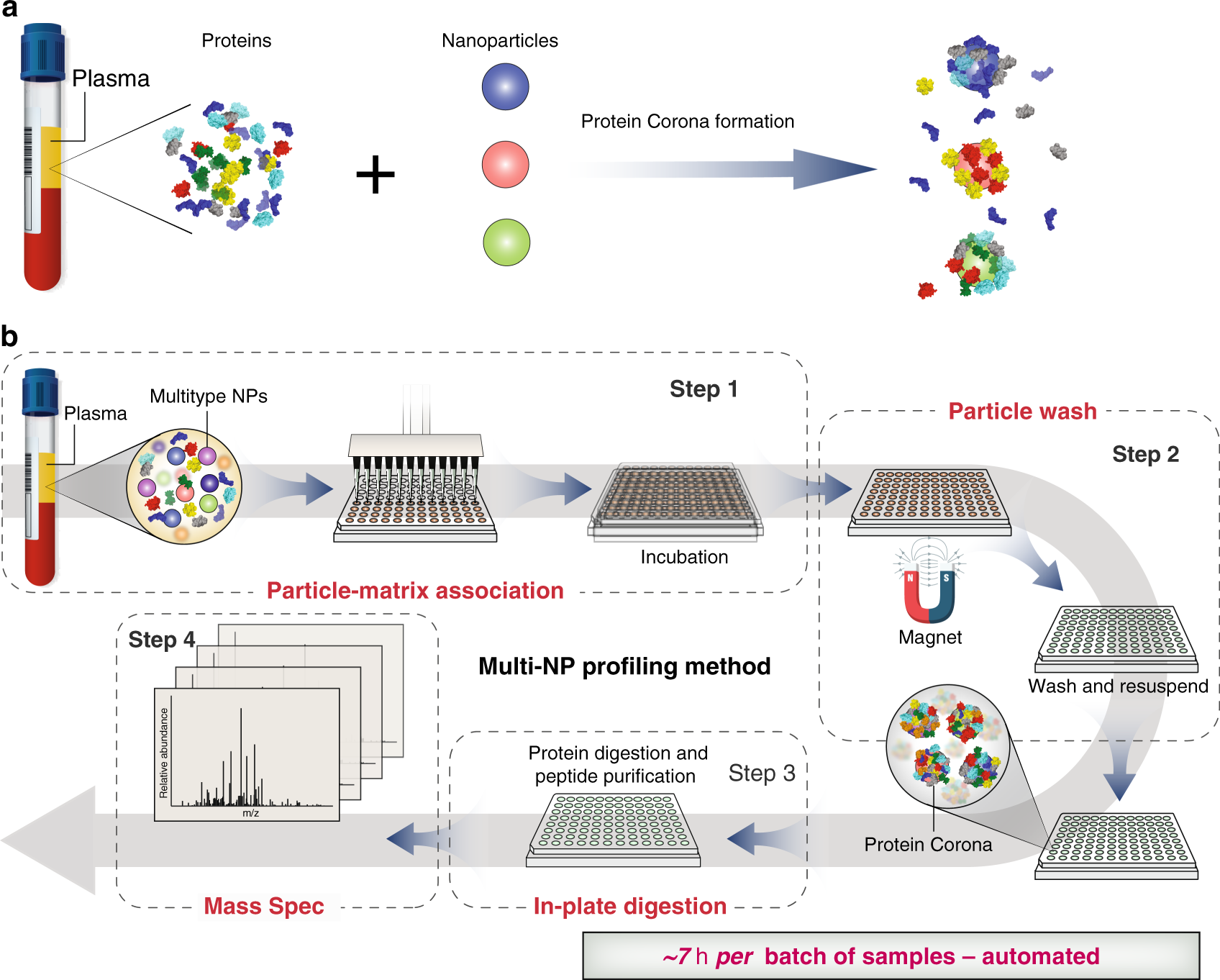
Rapid, deep and precise profiling of the plasma proteome with multi-nanoparticle protein corona | Nature Communications
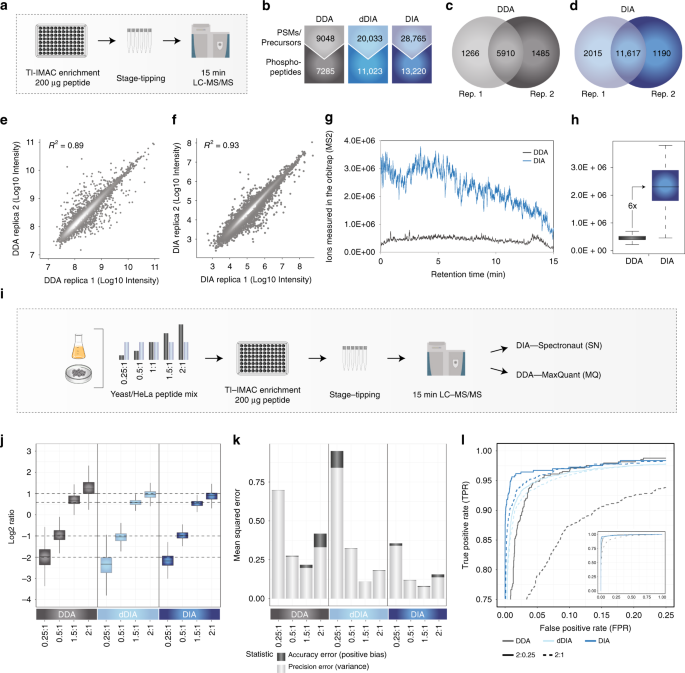
Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries | Nature Communications